This is the first article I have written after a one-year pause. So let's just start...
Cutting to the chase
There are articles on LCA on many sites(Topcoder, Codeforces), Youtube videos, the Wiki is quite explicative. So why do I write this? The answer is actually simple. I have no friends.
On a more serious note, I think most LCA articles don't mention the off-line and no-query cases.
The classic Lowest Common Ancestor
Take a tree with a fixed root. The Lowest Common Ancestor of two nodes x and y is the lowest(i.e. most far from the root) node s such that both x and y are descendants of the node s. For the following tree, we have:
LCA(4, 6) = 2
LCA(5, 10) = 1
LCA(6, 9) = 6
LCA(8, 9) = 2
LCA(4, 5) = 2
etc.
Suppose that we want to respond to multiple queries of type LCA(x, y) = ?. Assume you do not know all the queries in advance.
Let us start with the simple solution: just iterate through the ancestors of both x and y, keep the ancestors in two vectors and, finally, find the one that is the lowest one. This is awfully slow. A minor optimization is to actually stop at the LCA rather than at the root. Thus, we can calculate depth[x] = the distance from the root to node x. Now we can start with the node that has the maximum depth[x] and go the the ancestors until we reach depth[x] = depth[y]. Then we keep going with both the nodes to the ancestors until x = y. This will be the LCA.
This is nice, we used precomputed information to make this thing a bit more faster. Let us think a bit of what we did so far: we saw that going upper than the LCA was not necessary, so we filtered those nodes; in the second phase we considered the candidates at each level, until we found the right one. We basically found a way to filter out nodes that are not good to be our ancestor.
Let's make some observations regarding the l = LCA(x, y). Particularly, we want to: 1. distinguish l from its ancestors 2. distinguish l from other nodes on the path from x to y
We also want a different criteria to do this, as we want to combine those criteria into a single one.
For (1), the path between x and y passes through l and does not pass through any of the ancestors of l. An obvious choice for (2) is that depth[l] ≤ depth[k] for whatever k on the path from x to y.
The second criterion is a minimization problem, thus we think of using a data structure to get the minimum from a structure. This would be really a classic case if we had an array, thus we begin to think on how to flatten the tree. Addressing the first criterion, we need to flatten the tree such that between the positions of x and y we can find l and we can not find any ancestor of l.
Such a flattening is the Euler tour of the tree. The Euler tour of a tree is traversing the tree in a depth first form and adding the current node each time we arrive in it(i.e. even when we return from the recursion). The following picture is quite explicative.
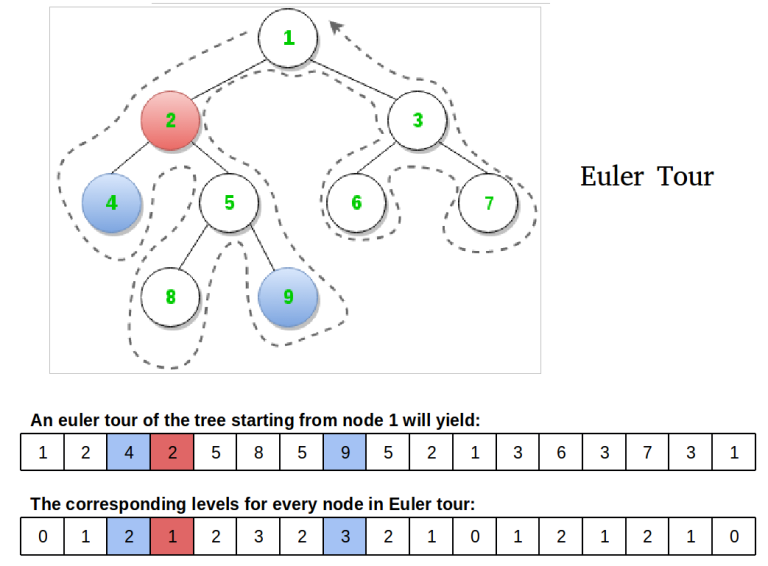
Now, let us see how to find the LCA of two nodes on this flattening. The LCA of node x and y is the array element with the minimum depth that is placed in the sequence between the first appearance of x and y. For nodes 4 and 5, we look at the sequence 4, 2, 5, 8, 5, 9. Node 2 has the smallest depth.
The final details is that we need a data structure that can compute the minimum from a substring fast.
Big Small Excursion
The problem we arrived at is: Given a fixed array a with n elements, and m queries of type [p1, p2], for each query [p1, p2] find min(a[p1], a[p1 + 1], ..., a[p2 + 1]). This is exactly the Range Minimum Query problem.
Let us start small again. Is is clear that we want to process stuff before the queries rather that using the naive solution. We observe that min(a[p1], ..., a[p2]) = max(max(a[p1], ..., a[p2 - 1]), a[p2]). This gives the idea of solving with DP: dp[p1][p2] = min(a[p1], ..., a[p2]), with the recurrence dp[p1][p2] = min(dp[p1][p2 - 1], a[p2]). This gives a O(n2 + m) solution.
The inefficiency in our solution seems to come from the fact that we do minimum of arrays (a[p1], a[p1 + 1], ..., a[p2 - 1]) and (a[p2]), rather than splitting equally the workload: (a[p1], a[p1 + 1], ..., a[k]) and (a[k + 1], a[k + 2], ..., a[p2]) where k = (p1 + p2) / 2.
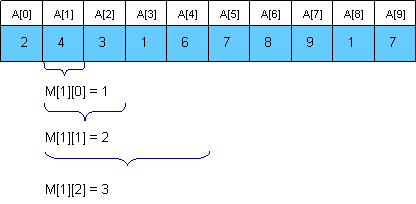
Starting from this idea and from the fact that overlapping two parts of our query does not affect us, let's define: dp[p][k] = min(a[p], a[p + 1], ..., a[p + 2k]). The recurrence will be: dp[p][k] = min(dp[p][k - 1], dp[p + 2(k - 1)][k - 1]). To respond to a query we (p1, p2), we have min(a[p1], ..., a[p2]) = min(dp[p1][k], dp[p2 - 2k + 1][k]) where k is the biggest number s.t. 2k ≤ p2 - p1 + 1. Refer to the stolen picture from Topcoder for details.
That gives a O(n·logn + m) solution as we can also precompute k. That's quite fast. That's what she said...
No-query case
Moving to the hipster stuff, let's look a bit at the simple case of LCA of two nodes. (this is also explained — I think quite similarly — in an older post of mine)
The obvious solution is to go upwards to the root and store the distances between the nodes(x and y) and the root. As long as dist(x, root) < dist(y, root), y will become f(y). When the distances are equal we are going upwards with both x and y. The complexity is O(R) , where R = max(dist(x, root), dist(y, root)).
A better solution comes with an observation. Take distance distance L = max(dist(x, LCA), dist(y, LCA)). Now if you go L nodes upwards from y, then L nodes upwards from x at some point in x’s upwards movement, you encounter the LCA. If you go a distance L’ ≥ L, then you end up meeting at a common ancestor of x and y, but not the lowest one. From here you can count the number of upward movements for each node and afterwards find the LCA.
The only problem now is how to find a value L’ ≥ L which has the same order of complexity as L. We can use binary search. We will do the algorithm described before with lengths l = 20, 21, 23, … until we will have the first l = 2k which will be larger than L. The complexity will be: O(1 + 2 + 4 + … + 2k) = O(2k + 1) = O(L).
A different idea is to alternate when we move , we apply the ascending function once for x, twice for y,4 times for x and so on. We have the guarantee that the roads will overlap at some moment. So we’ve managed to reduce the constant using that trick.
Off-line case
Off-line queries allow a different approach.
Given the fact that we attach the queries to the tree, we can keep the lowest common ancestor of partial trees that have been traversed so far. This forest of trees traversed so far can be imagined as a "wave" that propagates from left to right.
An algorithm based on this is Tarjan's off-line lowest common ancestors algorithm. The idea behind the algorithm is that we can process the queries from the leafs up to the root. I find the code very clean and clear. The set P is the set of queries, node u is the current node being traversed and the Union and Find operations are operations on disjoint data sets.
function TarjanOLCA(u)
MakeSet(u);
u.ancestor := u;
for each v in u.children do
TarjanOLCA(v);
Union(u,v);
Find(u).ancestor := u;
u.colour := black;
for each v such that {u,v} in P do
if v.colour == black
print "Tarjan's Lowest Common Ancestor of " + u +
" and " + v + " is " + Find(v).ancestor + ".";
A node is marked black if its subtree has been processed. You can see that the union between subtrees of node u(say v1 and v2) is made before the full tree has been processed. This is done so we can embed the queries in set P right on the tree. It is also important to note that the pair of nodes {u, v} is processed immediately after node u has been colored in black.
The algorithm's correctness can be proved by contradiction: suppose the ancestor found is not the lowest, then we can prove it should have been found on previous steps of the algorithm. The complexity is O((N + M)·log * N). Here is an implementation for reference.
PS --
Thank you for reading and please state your opinion on my tutorial. (or, more specifically, on my writing style and how useful you find the material presented) Any suggestions for next tutorial are welcome.
I found the off-line case quite cool when I found about it. Hope you enjoyed!






